My working hypotesis
"If before phiMU01 and phiMU02 prophages were active mycobacteriophages probably Phage integrase of D29 mycobacteri0phage (or L5) could have some common features with these prophages and with the prophage integrase inside the genome of Mycobacterium ulcerans ".
This analysis could give strength to my conviction about the presence of a new specific and unknown mycobateriophage usable in Buruli disease but at moment this phage has not been isolated in the soil or in fresch water or in other sources .
Mycobacteriophage D29
D29p32
D29p32
Integrase [DNA replication, recombination, and repair]; Region: XerC; COG0582
Phage integrase family; Region: Phage_integrase; pfam00589
Phage integrase, N-terminal SAM-like domain; Region: Phage_int_SAM_3; pfam14659
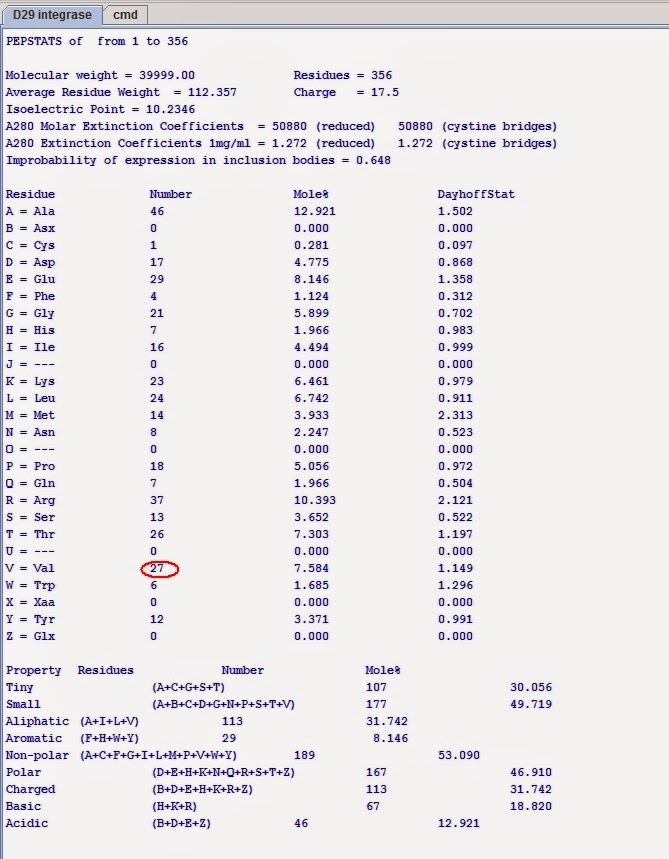
Statistics on the protein properties:
Distinguishing mark: VAL is present 27 times
Prophage integrase sequences from Mycobacterium ulcerans genome.
I have chosen MUL 0529:
MUL 0529
Site-specific recombinase XerC [DNA replication, recombination, and repair]; Region: XerC; COG4973
DNA breaking-rejoining enzymes, C-terminal catalytic domain. The DNA breaking-rejoining enzyme superfamily includes type IB topoisomerases and tyrosine recombinases that share the same fold in their catalytic domain containing six conserved active site...; Region: DNA_BRE_C; cl00213
Statistics on the protein properties:
Distinguishing mark: VAL is present 27 times
Comparison:
MUL 0529 Protein / D29p32 Protein Dot plot

D29p32 and MUL 0529 alignment

1-by Jemboss and by Wordcount software I select the word with 3 Aa but I chose only the words that contain Val Aa.
2- in the protein sequence I write all positions of VAL Aa,the start and the end of the main miscellaneous features.
3- in the protein sequence I check the positions of the words with the Val Aa.
In Position 91 in both the sequences is present VAL Aa .
In position 167 and 166 there is a VAL Aa.
LDV and TVP words are present in both the sequences and my working hypotesis is in part confirmed.
phiMU01 prophage
I have the confirmation of my working hypotesis.
By Swiss-Model there is also the confirmation:
The reference Model :
My contribution by Discovery Studio 4.0: